Cloning vectors are specialized plasmids (or other genetic elements) that will hold any piece of foreign DNA for further study or manipulation. The numbers and types of plasmids available for cloning have grown. In addition, other DNA elements are now used, including viruses and artificial chromosomes. Once a fragment of DNA has been cloned and inserted into a suitable vector, large amounts of DNA can be manufactured, the sequence can be determined, and any genes in the fragment can be expressed in other organisms. Studying human genes in humans is virtually impossible because of the ethical ramifications.
In contrast, studying a human gene expressed in bacteria provides useful information that can often be applied to humans. Modern biotechnology depends on the ability to express foreign genes in model organisms. Before discussing how a gene is cloned, the properties of vectors are considered.
Useful Traits for Cloning Vectors
Although many specialized vectors now exist, the following properties are convenient and found in most modern generalized cloning plasmids:
i) Small size, making them easy to manipulate once they are isolated
ii) Easy to transfer from cell to cell (usually by transformation)
iii) Easy to isolate from the host organism
iv) Easy to detect and select
Multiple copies helps in obtaining large amounts of DNA, Clustered restriction sites (polylinker) to allow insertion of cloned DNA
Method to detect presence of inserted DNA (e.g., alpha complementation)
Most bacterial plasmids satisfy the first three requirements. The next key trait of cloning vectors is an easy way to detect their presence in the host organism.
Bacterial cloning plasmids often have antibiotic resistance genes that make bacteria resistant to particular antibiotics. When treated with the antibiotic, only bacteria with the plasmid-borne resistance gene will survive. Other bacteria will die. Other traits have been exploited to detect plasmids.
Vectors derived from the yeast 2 µ plasmid often carry genes for essential amino acids, such as leucine. The host strain of yeast is defective in the corresponding gene and unable to grow on media lacking the amino acid, unless the plasmid is present.
Plasmids vary in their copy number . Some plasmids exist in just one or a few copies in their host cells whereas others exist in multiple copies. Such multicopy plasmids are in general more useful as the amount of plasmid DNA is higher, making them easier to isolate and purify.
The type of origin of replication controls the copy number, since this region on the plasmid determines how often DNA polymerase binds and induces replication. Most cloning vectors have several unique restriction enzyme sites. Usually these sites are grouped in one location called the polylinker or multiple cloning site. This allows researchers to open the cloning vector at one site without disrupting any of the vector’s replication genes.
Fragments of foreign DNA are digested with enzymes matching those in the polylinker. Ligase connects the vector and insert. Specific restriction enzyme sites can be added using PCR primers or synthetic DNA oligomers.
Some vectors have ways to detect whether or not they contain an insert. The simplest way to do this is insertional inactivation of an antibiotic gene.
Here, the vector has two different antibiotic resistance genes. The foreign DNA is inserted into one of the antibiotic-resistant genes. Thus the host bacteria will be resistant to one antibiotic and sensitive to the other.
Alternatively, alpha complementation may be used. The vector has a short portion of the β -galactosidase gene (the alpha fragment), and the bacterial chromosome has the rest of the gene. If both give rise to proteins, the subunits combine to form functional β -galactosidase.
If DNA is inserted into the plasmid-borne gene segment, the encoded subunit is not made and β -galactosidase is not produced. When β -galactosidase is expressed, the bacteria can degrade X-gal, which turns the bacterial colony blue. If a piece of DNA is inserted into the alpha fragment gene, the bacteria cannot split X-gal and stay white.
Once an appropriate vector has been chosen for the gene of interest or other insert, the two pieces are ligated into one construct. The term construct refers to any recombinant DNA molecule that has been assembled by genetic engineering.
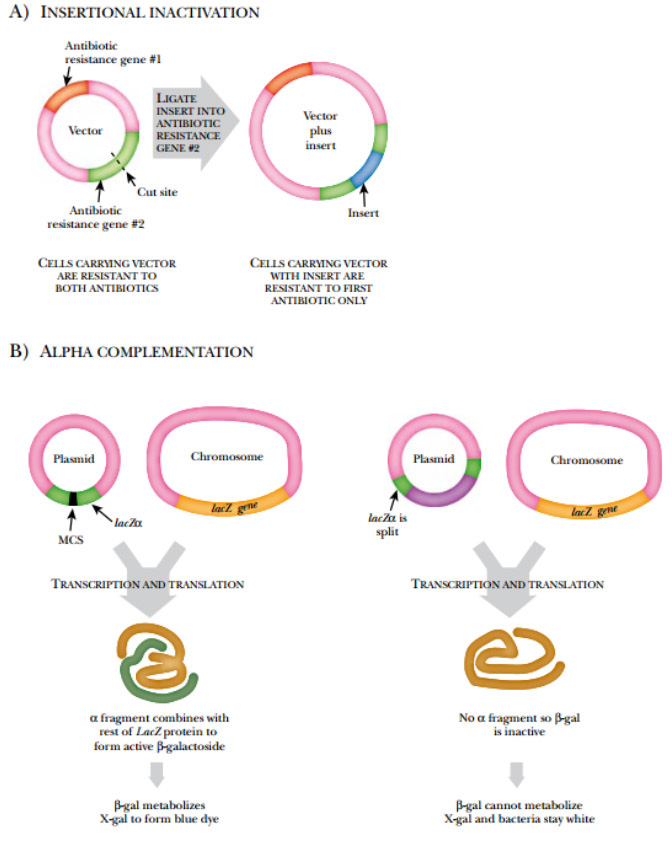
FIGURE: Detecting Inserts in Plasmids
(A) Insertional inactivation. Cells with an insert become sensitive to the second antibiotic. Cells without an insert remain resistant to the antibiotic. (B) Alpha complementation. Alpha complementation refers to the ability of β -galactosidase to be expressed as two protein fragments, which assemble to form a functional protein. In cells without an insert in the plasmid, β -galactosidase is active and splits X-gal to form a blue dye. In cells with an insert, the alpha fragment is not made and β -galactosidase is inactive. These cells remain white on media with X-gal.
If both the vector and insert are cut with the same restriction enzyme, the two pieces have complementary ends and require only ligase to link them. Tricks are used to make two pieces of DNA with unrelated ends compatible. Sometimes, short oligonucleotides are synthesized and added onto the ends of the insert to make them compatible with the vector. These short oligonucleotides are called linkers , and they add one or a few new restriction enzyme sites to the ends of a segment of DNA.