The first step in genetically engineering a plant is to identify a gene that will confer a specific desirable trait on the plant. In some ways, this is the hardest part of genetic engineering. The most desirable traits for a crop are increases in the amount of seed, grain, or other plant products. Increased resistance to disease or drought is also very useful. Finding the genes responsible is difficult, because multiple interacting genes usually control such traits. In addition, such genes may play other roles in plant physiology or development.
So far, most successful genetic engineering of plants has relied on inserting one or a few genes that supply simple, yet useful, properties. For example, resistance to the herbicide glyphosate is due to a single gene. Making a crop such as soybean resistant to glyphosate allows the farmer to kill the weeds in the field without harming the soybeans.
Another desirable trait often due to a single gene is the production of toxins that kill harmful insects.
Both these cases rely on transgenes derived from bacteria. As more research into plant physiology occurs, more genes can be identified that increase the value of a crop. For example, a two-gene pathway was engineered into rice to make it more resistant to drought
Plants can also be engineered for novel products. Thus, golden rice expresses the biosynthetic pathway for vitamin A precursors. This rice was developed for people who rely on rice as the one main food in their diet. The addition of vitamin A precursors can prevent deficiencies that cause blindness or premature death in children in developing countries. Researchers have also engineered the human insulin gene for expression in Arabidopsis and safflower. At present, insulin is produced by engineered bacteria.
However, plant-produced insulin is easier to isolate and purify in bulk and should cost much less. Further research will identify new genes and useful pathways that can be engineered into plants. Perhaps engineered plants will be used to clean up oil spills or other pollutants by growing them on contaminated soil.
GETTING GENES INTO PLANTS USING THE TI PLASMID
Plants suffer from tumors, though these are quite different from the cancers of animals. The most common cause is the Ti plasmid (tumor-inducing plasmid), which is carried by soil bacteria of the Agrobacterium group. Specifically, the Ti plasmid of Agrobacterium tumefaciens is an important tool for plant genetic engineering. The most important aspect of the infection is that a specific segment of the Ti plasmid DNA is transferred from the bacteria to the plant.
Scientists have exploited this genetic transfer in order to get genes with desired properties into plant cells. Agrobacterium is unique in the ability to transfer a segment of its DNA from one kingdom to another. Most DNA transfers occur only between closely related organisms.
In nature, Agrobacterium is attracted to plants that have minor wounds by phenolic compounds such as acetosyringone, which are released at the wound. These chemicals induce the bacteria to move and attach to the plant via a variety of cell surface receptors. The same inducers activate expression of the virulence genes on the Ti plasmid that are responsible for DNA transfer to the plant. This is under control of a two-component regulatory system At the cell surface, the sensor, VirA, is autophosphorylated when it detects the plant phenolic compounds.
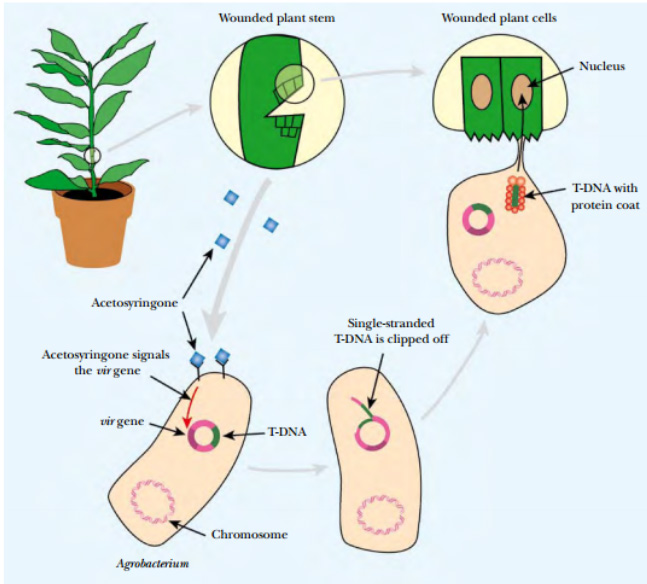
Fig: Agrobacterium Transfers Plasmid DNA into Infected Plants
Agrobacterium carrying a Ti plasmid is attracted by acetosyringone to a wounded plant stem. The Ti plasmid is cut by endonucleases to release single-stranded T-DNA, which is covered with protective proteins, and transported into the plant cell through a conjugation-like mechanism. The T-DNA enters the plant nucleus where it integrates into plant chromosomal DNA.
Next, VirA transfers the phosphate to the DNA-binding protein, VirG, which activates transcription of the vir genes of the Ti plasmid. Two of the gene products (VirD1 and VirD2) clip the T-DNA borders to form a single-stranded immature T-complex. VirD2 then attaches to the 5¢ end of the T-DNA, and bacterial helicases unwind the T-DNA from the plasmid. The single stranded gap on the plasmid is repaired, and the T-DNA is coated with VirE2 protein to give a hollow cylindrical filament with a coiled structure. This is the mature form of T-DNA and traverses into the plant.
T-DNA is transferred to the plant in a process similar to bacterial conjugation. First, Agrobacterium forms a pilus. This rodlike structure forms a connection with the plant cell and opens a channel through which the T-DNA is actively transported into the plant cytoplasm. Both pilus and transport complex consist of proteins that are vir gene products. Once inside the plant cytoplasm, T-DNA is imported into the nucleus. Both VirE2 and VirD2 have nuclear localization signals that are recognized by plant cytosolic proteins. These proteins take the T-complex to the nucleus where it is actively transported through a nuclear pore. The single T-DNA strand is integrated directly into the plant genome and converted to a double-stranded form. The integration requires DNA ligase, polymerase, and chromatin remodeling proteins, which are all supplied by the plant.
Once they are part of the plant genome, the genes in the T-DNA are expressed. These genes have eukaryote-like promoters, transcriptional enhancers, and poly(A) sites and hence are expressed in the plant nucleus rather than in the original bacterium.
The proteins they encode synthesize two plant hormones, auxin and cytokinin. Auxin makes plant cells grow bigger and cytokinin makes them divide. The infected plant cells begin to grow rapidly and without control, resulting in a tumor.
T-DNA also carries genes for synthesis of opines, which are a variety of different amino acid and sugar phosphate derivatives. The type of opine differentiates the various strains of Agrobacterium . Opines are made by plant cells that contain T-DNA but are used by the bacteria as carbon, nitrogen, and energy sources. Notice how the bacterium tricks the plant into using its resources to supply the bacteria with food. The Ti plasmid, which is still inside the Agrobacterium , carries genes that allow the bacteria to take up these opines and break them down for food. Note that other bacteria, which might be present by chance, cannot use opines because they do not possess the genes for uptake and metabolism. This ensures that the plant feeds only the bacteria with the Ti plasmid.
So how are Ti plasmids used to improve plants?
First, the Ti plasmid is disarmed by cutting out the genes in the T-DNA for plant hormone and opine synthesis.
Then, the transgene of interest, such as an insect toxin gene, is inserted into the T-DNA region of the Ti plasmid. The Ti plasmid is also streamlined by removing genes that are not involved in moving the T-DNA. These smaller plasmids are much easier to work with and can be manipulated in Escherichia coli rather than their original host, Agrobacterium.
Now, when the T-DNA enters the plant cell and integrates into the chromosome, it will bring in the transgene instead of causing a tumor.
The transferred region of the plasmid must also have other elements in order for the transgene to function properly. Expression of the transgene requires a promoter that works efficiently in plant cells. This may be one of two types. A constitutive promoter will turn the gene on in all the plant cells throughout development; thus every tissue, even the fruit or seed, will express the gene. A more refined approach is to use an inducible promoter that has an on/off switch. An example of this is the cab promoter from the gene encoding chlorophyll a / b binding protein.

FIGURE: Essential Elements for Carrying a Transgene on Ti Plasmids
The T-DNA segment contains both a transgene and a selective marker or reporter gene. These have separate promoters and termination signals. The marker or reporter gene must be expressed all the time, whereas the transgene is often expressed only in certain tissues or under certain circumstances and usually has a promoter that can be induced by appropriate signals.
This promoter is turned on only when the plant is exposed to light; therefore, root tissues and tubers such as potatoes will not express the gene. Many different promoters may be used, but ideally, the promoter should turn on only in tissues that need transgene function. Another important component for the genetically modified T-DNA region is some sort of selectable marker. Including an herbicide or antibiotic resistance gene in the T-DNA region can be used to track whether the foreign DNA has been inserted into plant cells. The selectable marker may cause problems because it must be expressed constitutively throughout the plant. Many people worry that the protein product of the selectable marker could cause allergies or reactions if expressed in fruit, grain, or vegetables. However, systems exists that can remove this gene once the transgenic plant has been isolated.
In practice, Agrobacterium is used to transfer genes of interest into plants using tissue culture. Either dissociated plant cells called protoplasts or a piece of callus are cultured with Agrobacterium harboring a Ti plasmid with modified T-DNA. After coculture, the plant cells are harvested and incubated with the herbicide or antibiotic used as the selectable marker. This kills all the cells that were not transformed with T-DNA or failed to express the genes on the T-DNA. The transformed cells can then be induced to produce shoot and root tissue by altering the hormone conditions in the medium as described earlier. The small transgenic plants can then be screened for transgene expression levels.
PLnta Agrobacterium transformation
Recently, a method for in planta Agrobacterium transformation was developed and has revolutionized the plant transformation world. In planta transformation is also known as the floral dip method. The method was developed using the model plant Arabidopsis but has been extended to other plants, such as wheat and maize.
First, Arabidopsis plants are grown until flower buds begin to form. These buds are removed and allowed to regenerate for a few days. Once they begin to regenerate, the plants are dipped into a suspension of Agrobacterium containing a surfactant. The surfactant allows the Agrobacterium to adhere to the plant and transfer its T-DNA.
Because the flower buds are just beginning to form, the T-DNA somehow becomes part of the germline through the ovarian tissue. The plant is allowed to finish growing and set seed. These seeds are harvested and grown in selective media to find those that have integrated and expressed T-DNA. Although the method gives a low percentage of transformants, so many seeds can be screened that the overall procedure works well.
PARTICLE BOMBARDMENT TECHNOLOGY