The ability to isolate, separate, and visualize DNA fragments would be useless unless some method was available to cut the DNA into fragments of different sizes. In fact, naturally occurring restriction enzymes or restriction endonucleases are the key to making DNA fragments. These bacterial enzymes bind to specific recognition sites on DNA and cut the backbone of both strands. They evolved to protect bacteria from foreign DNA, such as from viral invaders. The enzymes do not cut their own cell’s DNA because they are methylation sensitive, that is, if the recognition sequence is methylated, then the restriction enzyme cannot bind.
Bacteria produce modification enzymes that recognize the same sequence as the corresponding restriction enzyme. These methylate each recognition site in the bacterial genome. Therefore, the bacteria can make the restriction enzyme without endangering their own DNA.
Restriction enzymes have been exploited to cut DNA at specific sites, since each restriction enzyme has a particular recognition sequence. Differences in cleavage site determine the type of restriction enzyme.
Type I restriction enzymes cut the DNA strand 1000 or more base pairs from the recognition sequence.
Type II restriction enzymes cut in the middle of the recognition sequence and are the most useful for genetic engineering.
Type II restriction enzymes can either cut both strands of the double helix at the same point, leaving blunt ends , or they can cut at different sites on each strand leaving singlestranded ends, sometimes called sticky ends.
The recognition sequences of Type II restriction enzymes are usually inverted repeats, so that the enzyme cuts between the same bases on both strands. Since the repeats are inverted, the cuts may be staggered, thus generating single-stranded overhangs.
The number of base pairs in the recognition sequence determines the likelihood of cutting. Finding a particular sequence of four nucleotides is much more likely than finding a six base-pair recognition sequence.
So to generate fewer, longer fragments, restriction enzymes with six or more base-pair recognition sequences are used. Conversely, four base-pair enzymes give more, shorter fragments from the same original segment of DNA.
When two different DNA samples are cut with the same sticky-end restriction enzyme, all the fragments will have identical overhangs. This allows DNA fragments from two sources (e.g., two different organisms) to be linked together Fragments are linked or ligated using DNA ligase , the same enzyme that ligates the Okazaki fragments during replication. The most common ligase used is actually from T4 bacteriophage. Ligase catalyzes linkage between the 3′-OH of one strand and the 5′-PO 4 of the other DNA strand. Ligase is much more efficient with overhanging sticky ends, but can also link blunt ends much more slowly. (Specific fragments for ligation are often isolated by agarose gel electrophoresis as described earlier.)
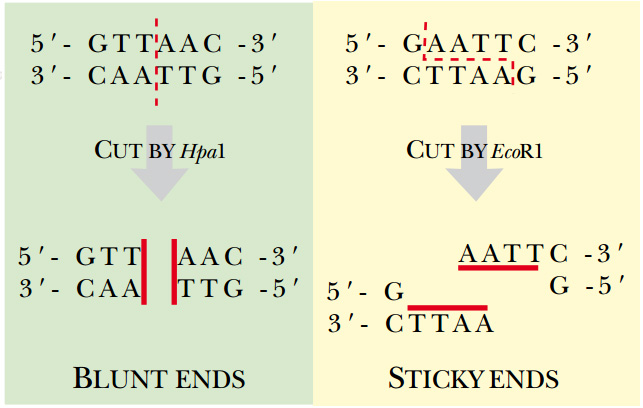
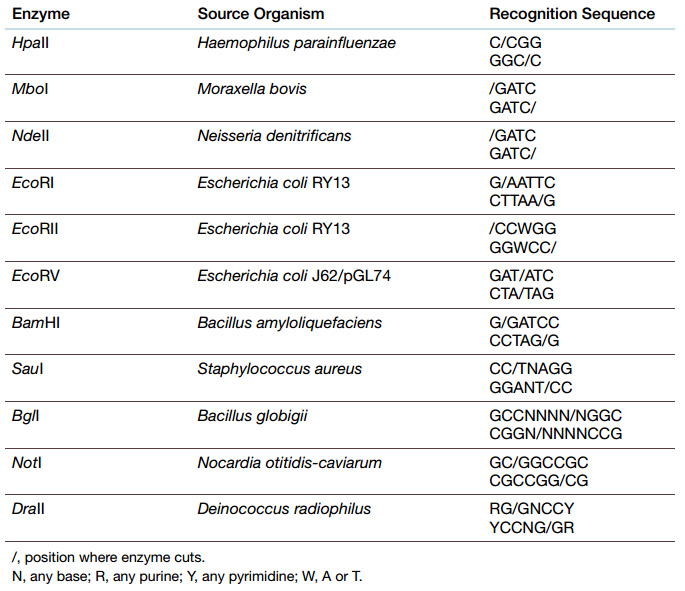
Fig: Type II Restriction Enzymes—Blunt versus Sticky Ends Hpa I is a blunt end restriction enzyme, that is, it cuts both strands of DNA in exactly the same position. Eco RI is a sticky end estriction enzyme. The enzyme cuts between the G and A on both strands, which generates four basepair overhangs on the ends of the DNA. Since these ends may base pair with complementary sequences, they are considered “sticky.”
Restriction Fragment Length Polymorphisms Identify Individuals
Restriction enzymes are useful for many different applications. Because the DNA sequence is different in each organism, the pattern of restriction sites will also be different. The source of isolated DNA can be identified by this pattern. If genomic DNA is isolated from one organism and cut with one particular restriction enzyme, a specific set of fragments can be separated and identified by electrophoresis. If DNA from a different organism is cut by the same restriction enzyme, a different set of fragments will be generated. This technique can be applied to DNA from two individuals from the same species. Although the DNA sequence differences will be small, restriction enzymes can be used to identify these differences. If the sequence difference falls in a restriction enzyme recognition site, it gives a restriction fragment length polymorphism (RFLP) ( Fig. A ). When the restriction enzyme patterns are compared, the number and size of one or two fragments will be affected for each base difference that affects a cut site.
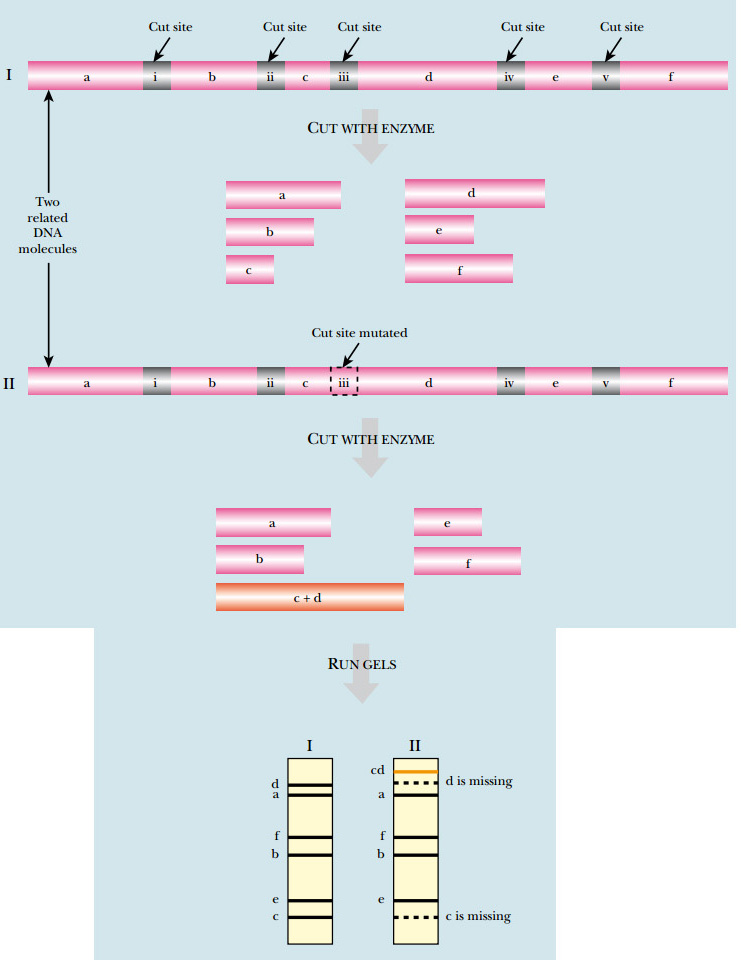
FIG: RFLP Analysis
DNA from related organisms shows small differences in sequence that cause changes in restriction sites. In the example shown, cutting a segment of DNA from the first organism yields six fragments of different sizes (labeled a – f on the gel). If the equivalent region of DNA from a related organism were digested with the same enzyme, a similar pattern would be expected. Here a single-nucleotide difference is present, which eliminates one of the restriction sites. Consequently, digesting this DNA produces only five fragments. Fragments c and d are no longer seen, but form a new band labeled cd .