The process of protein synthesis.
Charging of tRNA:
Each amino acid is attached to a tRNA molecule, specific to that amino acid by a high-energy bond derived from ATP. The process is catalyzed by a specific enzyme called a synthetase (the tRNA is called charged when amino acid is attached)
AA1 + tRNA1 + ATP à aa1-tRNA1 + ADP
(A) INITIATION:
The main task of initiation is to place the first aminoacyl-tRNA in the P site of the ribosome and, in this way, establish the correct reading frame of the mRNA.
In most prokaryotes and all eukaryotes, the first amino acid in any newly synthesized polypeptide is methionine, specified by the codon AUG.
It is inserted not by tRNAMet but by a special tRNA called an initiator, symbolized tRNAMeti.
In bacteria, a formyl group is added to the methionine while the amino acid is attached to the initiator, forming N-formylmethionine. (The formyl group on N-formylmethionine is removed later.)
How does the translation machinery know where to begin?
In both prokaryotes and eukaryotes, mRNA has a 5′ untranslated region consisting of the sequence between the transcriptional start site and the translational start site.
Shine–Dalgarno sequences
In prokaryotes, initiation codons are preceded by special sequences called Shine–Dalgarno sequences that pair with the 3′ end of an rRNA, called the 16S rRNA, in the 30S ribosomal subunit. This pairing correctly positions the initiator codon in the P site where the initiator tRNA will bind.
The mRNA can pair only with a 30S subunit that is dissociated from the rest of the ribosome. Note again that rRNA performs the key function in ensuring that the ribosome is at the right place to start translation.
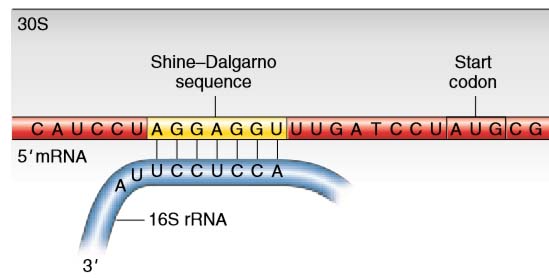
Figure 1 Shine–Dalgarno sequence. In bacteria, base complementarity between the 3′ end of the 16S rRNA of the small ribosomal subunit and the Shine–Dalgarno sequence of the mRNA positions the ribosome to correctly initiate translation at the downstream AUG codon.
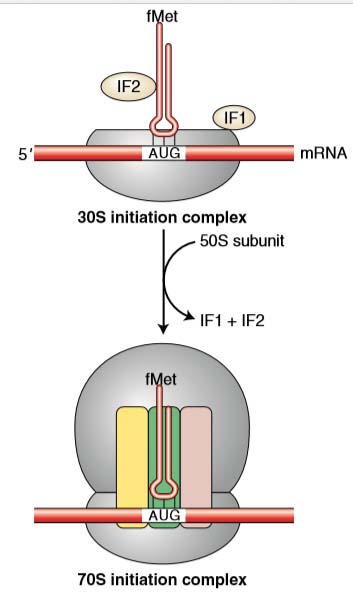
The first step in initiation involves binding of the mRNA to the 30 S subunit of ribosome. Initiation factor 3 (IF-3) controls this binding. The first codon AUG is exposed.
- IF2 binds to GTP and to the fMet-tRNAi, and stimulates the binding of fMet-tRNA to the initiation complex, leading to the fMet-tRNAi into the P site (peptidyl site).
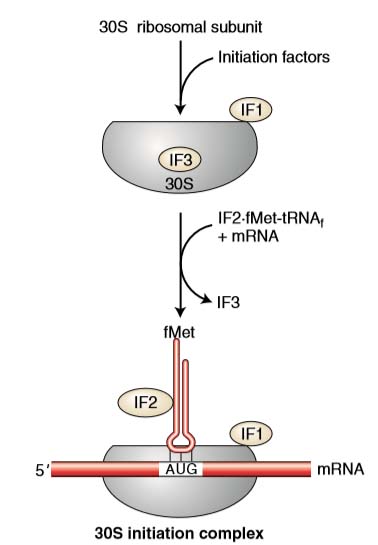
- Three regions are important as the ribosome is assembled around the mRNA. They are commonly called the A, P, and E sites.
- Each site will fit a single tRNA.
ELONGATION
- It is the assembly of specified amino acids into a polypeptide chain.
- Following initiation, the first tRNA (for methionine) is located within the P site.
- A second codon in the mRNA is exposed in the A site.
- Only a tRNA with an anticodon complementary to the mRNA codon exposed in the A site will correctly fit.
- At this point there are two tRNAs in the ribosome. By an enzymatic reaction, the amino acids between the P and A sites are joined together by a peptide bond.
- As the peptide bond forms, the amino acid is released from the tRNA in the P site. The ribosome then moves one codon down the mRNA (in the 3′ direction).
- As it does so, the tRNA that was in the P site enters into the E site and leaves the ribosome.
- The tRNA that was in the A site, which still has the polypeptide chain attached, moves into the P site.
- A new mRNA codon is then revealed in the A site.
- A tRNA with an anticodon complementary to the exposed mRNA codon then enters the A site, and the process repeats itself.
The rate at which the ribosome may read codon is up to six codons per second
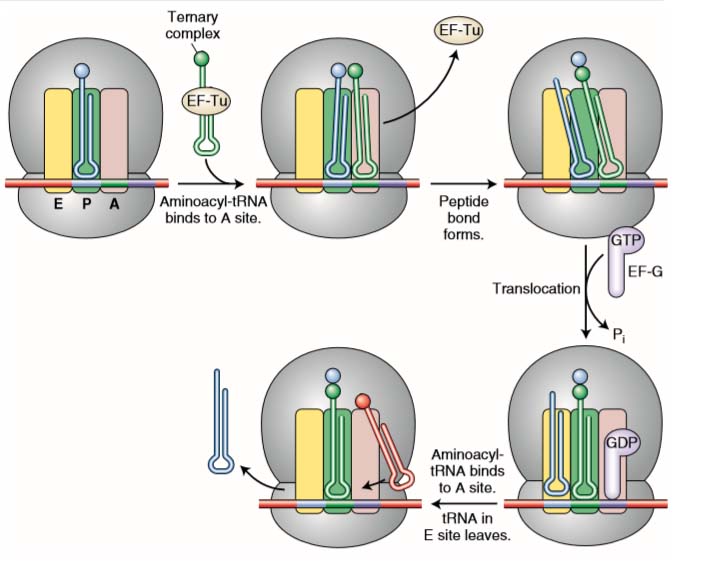
Figure 2 Steps in elongation. A ternary complex consisting of an aminoacyl-tRNA attached to an EF-Tu factor binds to the A site. When its amino acid has joined the growing polypeptide chain, an EF-G factor binds to the A site while nudging the tRNAs and their mRNA codons into the E and P sites. See text for details.
TERMINATION
When ribosome reaches at the end of mRNA one of the termination codon (UAA, UGA, UAG) exposed.
In place of tRNAs, proteins called release factors (RF1, RF2, and RF3 in bacteria) enter into the A site.
In bacteria, RF1 recognizes UAA or UAG, whereas RF2 recognizes UAA or UGA; both are assisted by RF3.
Release factors fit into the A site of the 30S subunit but do not participate in peptide-bond formation. Instead, a water molecule gets into the peptidyltransferase center and leads to the release of the polypeptide from the tRNA in the P site.
Since the release factors do not contain amino acids, the process of translation is stopped at this point. The ribosomes disassembled.
The polypeptide chain that is formed must undergoes a series of folds in order to produce a functional protein.

Figure 4 Termination of translation. Translation is terminated when release factors recognize stop codons in the A site of the ribosome