The process of doubling of DNA is called replication

Figure 1 DNA replication at the growing fork.
DNA REPLICATION IN PROKARYOTES
Origin site:
Replication starts at a single origin in the circular bacterial chromosome. The double helix opens up and both strands serve as template for the synthesis of new DNA.
Gyrase:
it is a type of topoisomerases and responsible for removing the supercoiling the DNA.
DNA helicase:
is responsible for separation of double stranded DNA into single stranded DNA.
Single-strand binding (SSB) protein
these attach with the single stranded regions of the DNA, and prevents it from recoiling.
DNA polymerase III (Kornberg enzyme, after Thomas Kornberg; 1970)
- It is a true replicating enzyme, which is 10 times larger and far more complex in structure pol-I and II.
- It is a dimmer, which catalyzes the process of replication.
- It adds nucleotides at the rate of1000 nucleotides / second from 5 to 3 direction only.
DNA Primase:
DNA polymerase-III only adds nucleotides to already present 3 end hydroxyl group. So the primer (a short sequence of RNA approximately 10-12 nucleotides), is synthesized by DNA primase.
Replication is bidirectional:
Double stranded DNA is antiparallel, one strand runs 5’- 3’, and the complementary strand runs 3’- 5’. As the original double stranded DNA opens up at a replication fork, new DNA is made against each template strand. Superficially, therefore, one might expect new DNA to be made 5’ 3’ for one daughter strand and 3’ 5’ for the other daughter strand. However, all DNA polymerases make DNA only in the 5’ 3’ direction and never with 3’ 5’ direction.
Leading strand:
On the template strand from 3’ 5’, new DNA synthesis is in a continuous piece in 5’ 3’ direction. This new DNA is called leading strand. (It elongates towards the replication fork)
Lagging Strand:
On the other template strand, DNA polymerase synthesize short pieces of new DNA (about 1000 nucleotides long) in the 5’ 3’ direction and then joins these pieces together. The small fragments are called Okazaki fragments, after the name of their discoverer. The new DNA strands which is made by this discontinuous method is called lagging strand (it elongates away from the replication fork)
DNA polymerase I (Kornberg enzyme, after Arthur Kornberg; 1956)
uses its 5’ 3’ exonuclease activity to remove the RNA primer and then fills the gap with new DNA.
DNA ligase
joins the ends of the DNA fragments together.
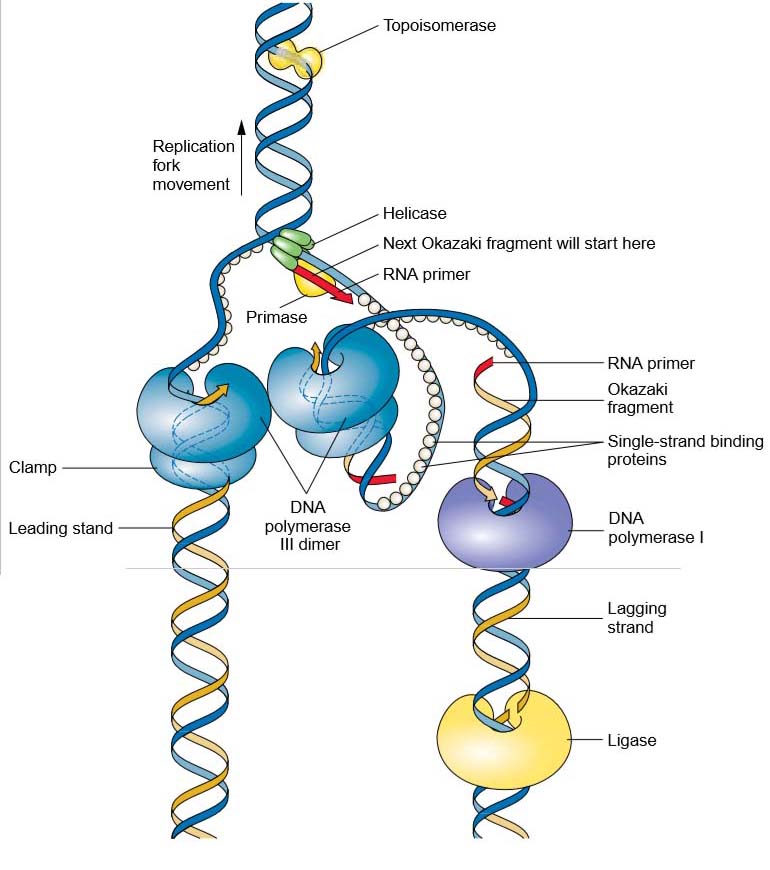
Figure 2 Details of the replisome and accessory proteins at the replication fork. Topoisomerase and helicase unwind and open the double helix in preparation for DNA replication. Once unwound, single-strand binding proteins prevent the double helix from reforming. The figure is a representation of the so-called trombone model (named for its resemblance to a trombone owing to the looping of the lagging strand) showing how the two catalytic cores of the replisome are envisioned to interact to coordinate the numerous events of leadingand lagging-strand replication.