DNA replication is more complex in eukaryotes in large part because there is a lot more DNA to copy. Transcription is more complicated in eukaryotes for following reasons.
- The larger eukaryotic genomes have many more genes to be recognized and transcribed. Whereas bacteria usually have a few thousand genes, eukaryotes have tens of thousands of genes.
- There is much more noncoding DNA in eukaryotes.
- So,even though eukaryotes have more genes than prokaryotes do,their genes are,on average,farther apart. For example,whereas the gene density (average number of genes per length of DNA) in
E. coli is 900 genes per million base pairs,
that number drops to 110 genes per million base pairs for the fruit fly Drosophila,
and it is only 9 genes per million base pairs for humans.
Eukaryotes deal with this situation in several ways.
4. They require the assembly of many proteins at a promoter before RNA polymerase II can begin to synthesize RNA. Some of these proteins,called general trancription factors (GTFs), bind before RNA polymerase II binds,whereas others bind afterward.
5. RNA is synthesized in the nucleus where the DNA is located and must be modified in several ways before it can be exported out of the nucleus into the cytoplasm for translation.
These modifications are collectively referred to as RNA processing.
First,they have divided the job of transcription among three different polymerases.
a. RNA polymerase I:
transcribes rRNA genes (excluding 5S rRNA).
b. RNA polymerase II
transcribes all protein-coding genes,for which the ultimate transcript is mRNA, and transcribes some snRNAs.
c. RNA polymerase III
transcribes the small functional RNA genes (such as the genes for tRNA,some snRNAs,and 5S rRNA).
Initiation
The core of RNA polymerase II also cannot recognize promoter sequences on its own. However, unlike the case for bacteria, where sigma factor is an integral part of the polymerase holoenzyme, GTFs are required in eukaryotes to bind to regions in the promoter before the binding of the core enzyme.
Preinitiation complex (PIC)
The GTFs and the RNA polymerase II core constitute the preinitiation complex (PIC).
This complex is quite large: it contains six GTFs, each of which is a multiprotein complex, plus the RNA polymerase II core, which is made up a dozen or more protein subunits.
Promoter sequence
Like prokaryotic promoters, eukaryotic promoters are located on the 5′ side (upstream) of the transcription start site.
TATA Box:
In an alignment of eukaryotic promoter regions, it can be seen that the sequence TATA is often located about 30 base pairs (-30 bp) from the transcription start site. This sequence, called the TATA box, is the site of the first event in transcription: the binding of the TATA binding protein (TBP).
When bound to the TATA box, TBP attracts other GTFs and the RNA polymerase II core to the promoter, thus forming the preinitiation complex.
After transcription has been initiated, RNA polymerase II dissociates from most of the GTFs to elongate the primary RNA transcript.
Some of the GTFs remain at the promoter to attract the next RNA polymerase core.
In this way, multiple RNA polymerase II enzymes can be synthesizing transcripts from a single gene at one time.
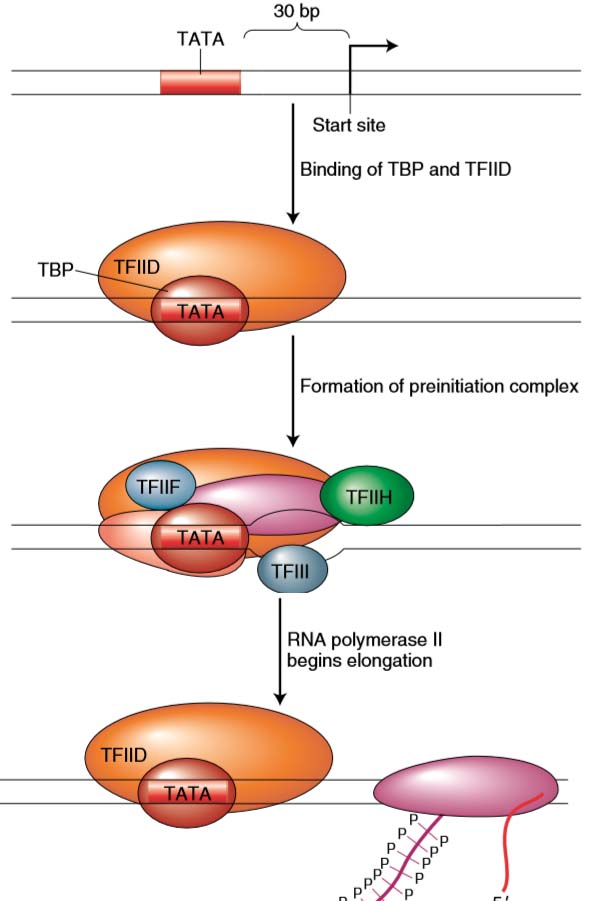
Figure Transcription initiation in eukaryotes. Formation of the preinitiation complex usually begins with the binding of the TATA binding protein (TBP), which then recruits the other general transcription factors and RNA polymerase II to the transcription start site. Transcription begins after phosphorylation of the carboxyl tail domain (CTD) of RNA polymerase II.
Processing 5′ and 3′ ends
the processing of the 5′ end of the transcript of a proteincoding gene.
CAP
When the nascent RNA first emerges from RNA polymerase II, a special structure, called a cap, is added to the 5. end by several proteins that interact with the CTD. The cap consists of a 7-methylguanosine residue linked to the transcript by three phosphate groups.
The cap has two functions.
First, it protects the RNA from degradation—an important step considering that a eukaryotic mRNA has a long journey before being translated.
Second, the cap is required for translation of the mRNA.
Tail
RNA elongation continues until the conserved sequence, AAUAAA or AUUAAA, near the 3′ end is recognized by an enzyme that cuts off the end of the RNA approximately 20 bases farther down. To this cut end, a stretch of 150 to 200 adenine nucleotides called a poly(A) tail is added.
Hence the AAUAAA sequence of protein-coding genes is called a polyadenylation signal.
Figure